Articles
- Page Path
- HOME > J Liver Cancer > Volume 22(2); 2022 > Article
-
Original Article
The diagnostic value of circulating tumor DNA in hepatitis B virus induced hepatocellular carcinoma: a systematic review and meta-analysis -
Young Chang1
 , Soung Won Jeong1
, Soung Won Jeong1 , Jae Young Jang1
, Jae Young Jang1 , Hyuksoo Eun2
, Hyuksoo Eun2 , Young‑Sun Lee3
, Young‑Sun Lee3 , Do Seon Song4
, Do Seon Song4 , Su Jong Yu5
, Su Jong Yu5 , Sae Hwan Lee6
, Sae Hwan Lee6 , Won Kim7
, Won Kim7 , Hyun Woong Lee8
, Hyun Woong Lee8 , Sang Gyune Kim9
, Sang Gyune Kim9 , Seongho Ryu10
, Seongho Ryu10 , Suyeon Park11,12
, Suyeon Park11,12
-
Journal of Liver Cancer 2022;22(2):167-177.
DOI: https://doi.org/10.17998/jlc.2022.09.19
Published online: September 29, 2022
1Institute for Digestive Research, Digestive Disease Center, Department of Internal Medicine, Soonchunhyang University College of Medicine, Seoul, Korea
2Department of Internal Medicine, College of Medicine, Chungnam National University, Daejeon, Korea
3Department of Internal Medicine, Guro Hospital, Korea University College of Medicine, Seoul, Korea
4Department of Internal Medicine, St. Vincent’s Hospital, College of Medicine, The Catholic University of Korea, Suwon, Korea
5Department of Internal Medicine and Liver Research Institute, Seoul National University College of Medicine, Seoul, Korea
6Department of Internal Medicine, Soonchunhyang University College of Medicine, Cheonan, Korea
7Department of Internal Medicine, Seoul Metropolitan Government Seoul National University Boramae Medical Center, Seoul National University College of Medicine, Seoul, Korea
8Department of Internal Medicine, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
9Department of Internal Medicine, Soonchunhyang University College of Medicine, Bucheon, Korea
10Soonchunhyang Institute of Medi-bio Science (SIMS), Soonchunhyang University, Cheonan, Korea
11Department of Biostatistics, Soonchunhyang University Hospital, Seoul, Korea
12Department of Applied Statistics, Chung-Ang University, Seoul, Korea
-
Corresponding author: Soung Won Jeong Institute for Digestive Research, Digestive Disease Center, Department of Internal Medicine, Soonchunhyang University Seoul Hospital, Soonchunhyang University College of Medicine, 59 Daesagwan-ro, Yongsan-gu, Seoul 04401, Korea
Tel. +82-2-710-3076, Fax. +82-2-709-9696 E-mail: jeongsw@schmc.ac.kr
Copyright © 2022 The Korean Liver Cancer Association
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- 2,923 Views
- 83 Downloads
- 2 Citations
Abstract
-
Background/Aim
- New biomarkers are urgently needed to aid in the diagnosis of early stage hepatocellular carcinoma (HCC). We performed a meta-analysis on the diagnostic utility of circulating tumor DNA (ctDNA) levels in patients with hepatitis B virus-induced HCC.
-
Methods
- We retrieved relevant articles from PubMed, Embase, and the Cochrane Library up to February 8, 2022. Two subgroups were defined; one subset of studies analyzed the ctDNA methylation status, and the other subset combined tumor markers and ctDNA assays. Pooled sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under the summary receiver operating characteristic curve (AUC) were analyzed.
-
Results
- Nine articles including 2,161 participants were included. The overall SEN and SPE were 0.705 (95% confidence interval [CI], 0.629-0.771) and 0.833 (95% CI, 0.769-0.882), respectively. The DOR, PLR, and NLR were 11.759 (95% CI, 7.982-17.322), 4.285 (95% CI, 3.098-5.925), and 0.336 (0.301-0.366), respectively. The ctDNA assay subset exhibited an AUC of 0.835. The AUC of the combined tumor marker and ctDNA assay was 0.848, with an SEN of 0.761 (95% CI, 0.659-0.839) and an SPE of 0.828 (95% CI, 0.692-0.911).
-
Conclusions
- Circulating tumor DNA has promising diagnostic potential for HCC. It can serve as an auxiliary tool for HCC screening and detection, especially when combined with tumor markers.
- In 2020, primary liver cancer was the sixth most common cancer and third leading cause of cancer-related deaths.1 Hepatocellular carcinoma (HCC) is the most common primary liver cancer (75-90% of all the cases).2 Of the several risk factors, chronic hepatitis B virus (HBV) infection is the most important, being associated with approximately 54% of HCC cases worldwide3 and 62.2% of HCC cases in Korea.4 Although early diagnosis is of utmost importance, HCC is often initially asymptomatic; most patients are diagnosed in the middle or late stages. Therefore, surveillance of patients with high-risk features on ultrasonography (US), with or without the measurement of α-fetoprotein (AFP) levels, is important.5,6 However, the recommended surveillance methods vary between studies. The sensitivity (SEN) of US-mediated early stage HCC detection in high-risk patients (with and without serum AFP measurement) was approximately 60%.7-9 New biomarkers that facilitate early detection are thus required. Circulating tumor DNA (ctDNA) fragment assays (also known as ‘liquid biopsies’) may aid HCC detection10,11 because ctDNA contains all of the genetic information of tumor cells and can be detected in the peripheral blood of early stage HCC patients.12,13 HCC development is accompanied by genetic and epigenetic mutations in the ctDNA. HCC-specific mutations in the plasma DNA have been reported in several studies.14-16 However, given the low amounts of ctDNA in patients with early stage cancers, it is difficult to distinguish true mutations from PCR and sequencing errors.17 A panel of common HCC-associated mutations has been used to diagnose HCC. Genomic profiling has identified key driver mutations in TP53, CTNNB1, PIK-3CA, PTEN, AXIN1, the promoter of TERT, and integrated HBV genomes.18-20 Both hyper and hypomethylated ctDNA regions are used for early tumor detection, and epigenetic changes play important roles in carcinogenesis.21,22 It is easier to detect, amplify, and quantify aberrant ctDNA hypermethylation than to document genomic mutations.23 In humans, many 5-hydroxymethylcytosine (5hmC) epigenetic markers are generated via oxidation of 5-methylcytosine by ten-eleven translocation enzymes.24 Such biomarkers aid in precision medicine because they reflect cancer gene regulation at the tissue level. Technical advances have made liquid biopsies convenient.25-28 However, although many studies have reported that HCC ctDNA assays are diagnostic, they vary extensively in terms of design and results. Therefore, there is an urgent need to assess the diagnostic utility of HCC ctDNA assays. Since the etiology of HCC could be one of the reasons for the heterogeneity among previous studies, we performed a systematic literature review and meta-analysis to assess the diagnostic utility of ctDNA assays in HBV-induced HCC.
INTRODUCTION
- 1. Search strategy and literature selection
- The meta-analysis followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Supplementary Table 1).29 We systematically searched PubMed, Embase, and the Cochrane library. The language was limited to English and the search was limited to humans. The search terms were “circulating tumor DNA” OR “circulating DNA” OR “ctDNA” OR “plasma DNA” OR “serum DNA” OR “liquid biopsy” OR “cell free tumor DNA” OR “cell free nucleic acid” OR “circulating cell free nucleic acid” OR “circulating nucleic acid” OR “cell free DNA” AND “liver cancer” OR “hepatocellular carcinoma” OR “liver neoplasm” OR “hepatic carcinoma” OR “liver tumor” OR “liver cell carcinoma” OR “hepatocarcinoma” OR “hepatoma” OR “liver cell cancer” OR “hepatic cancer” OR “hepatic neoplasm” OR “hepatocellular cancer” OR “hepatocellular neoplasm” OR “hepatocellular tumor” OR “liver cell neoplasm” OR “HCC” AND “chronic hepatitis B” OR “chronic hepatitis B virus infection” OR “hepatitis B virus” OR “chronic HBV” OR “HBV infection” OR “hepatitis B infection.” We manually reviewed the reference lists and added the relevant articles.
- 2. Inclusion and exclusion criteria
- The inclusion criteria were HBV-induced HCC, HCC ctDNA assay of serum or plasma, use of ctDNA data for initial HCC diagnosis, and availability of sensitivity (SEN) and specificity (SPE) data (either explicit or calculable). The exclusion criteria were reviews, cases, abstracts, letters, comments, meta-analyses, duplicate reports, predictions of HCC risk or prognosis, no SEN or SPE data, and/or analysis of ctDNAs that are not in the serum or plasma (rather in the liver or urine).
- 3. Data extraction and quality assessment
- Two investigators independently selected relevant studies and summarized the results. We collected the following data: first author, publication year, references, study type, control modality, sample size, sampling time, sample source, detection methods, assay indicators, cutoff values, SEN, SPE, true positive (TP), true negative (TN), false positive (FP), and false negative (FN) values. Both investigators independently assessed the study quality using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS)-2 criteria30 and any disagreement was resolved via discussion.
- 4. Statistical analysis
- The TP, TN, FP, and FN data were extracted from all the studies. Heterogeneity was assessed by calculating the I2 statistic. An I2 value >50% reflected significant heterogeneity, and a random-effects model was then used for the analysis. Threshold effects, regression, and subgroup analyses were used to identify sources of heterogeneity. We derived the pooled SEN, SPE, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), summary receiver operating characteristic curve (SROC), and area under the curve (AUC) for all subjects and subgroups. The Fagan Nomogram was used to validate clinical utility and the Deek funnel plot asymmetry test was used to detect publication bias (P<0.10 indicated such bias). We utilized the Rex software (version 3.6.0, Rex Soft Inc., Seoul, Korea) for analysis.
METHODS
- 1. Study characteristics
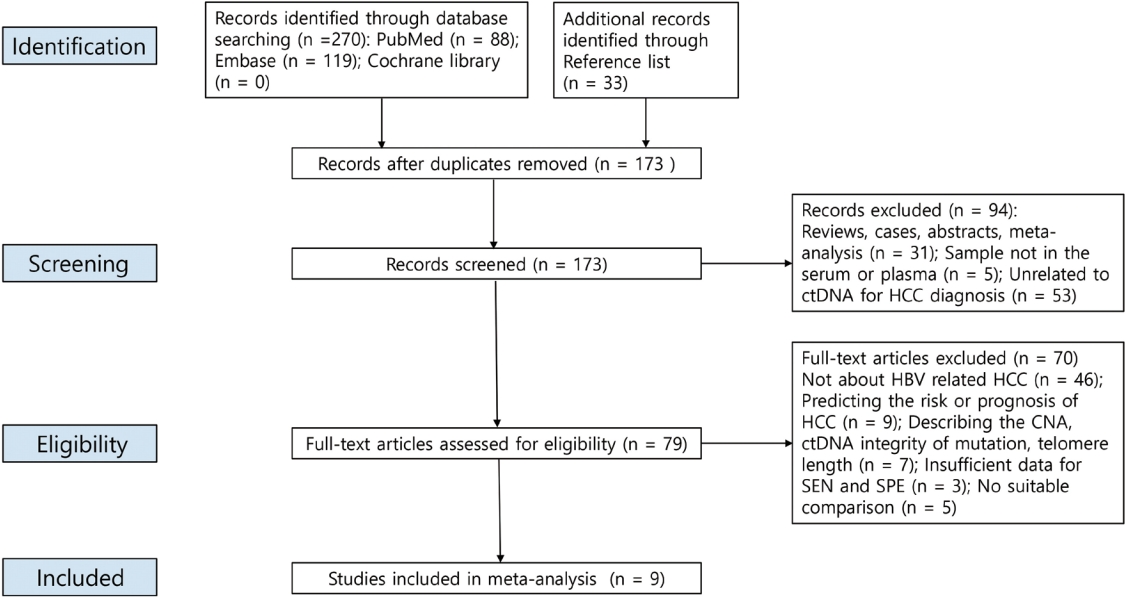
- The PRISMA flow diagram is shown in Fig. 1. We initially retrieved 303 publications, but ultimately included nine studies19,31-38 from 2013 to 2019 after excluding duplicates and studies that were poorly designed, and carefully reviewed the titles, abstracts, and full texts. The included studies enrolled 1,042 HCC patients and 1,119 controls. Most of the controls had benign liver disease (chronic hepatitis B or cirrhosis), but 149 of them were healthy. Table 1 summarizes the significant features of these studies. Of the nine studies, seven explored the diagnostic utility of ctDNA methylation32-38 and two explored the utility of other genetic variants in HCC ctDNA.19,31 Seven articles assessed the diagnostic utility of a combination of ctDNA and tumor marker assays.19,32-37 In the six studies that reported sampling times, five samples were collected before treatment, and one sample was collected before diagnosis. ctDNA was obtained from the plasma (n=2) or serum (n=7). The assay methods used included methylation-specific polymerase chain reaction and bisulfite sequencing.
- 2. Quality assessment
- The quality of the nine studies is shown in Fig. 2 and Supplementary Table 2. Most were of moderate-to-high quality. However, four patients were at risk of patient selection bias because consecutive or random sampling was absent, the study design was not case-controlled, or patients were inappropriately excluded. Four studies may be associated with an unknown risk of bias in terms of the index test; it is unclear if the test results were interpreted without knowledge of the reference data or whether the threshold was predefined. All the reference standards exhibited a low risk of bias.
- 3. Diagnostic value of ctDNA assay in HBVinduced HCC patients
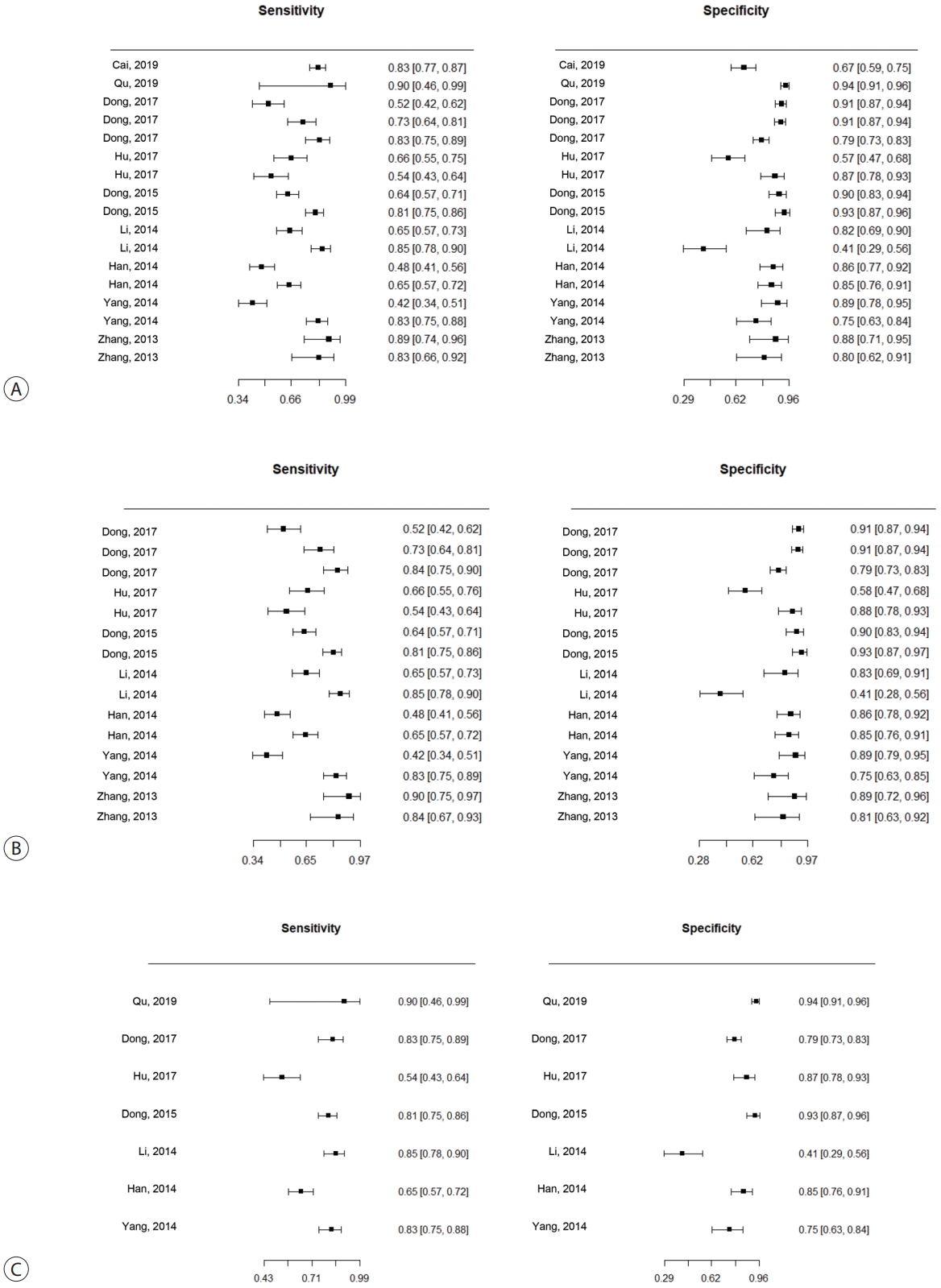
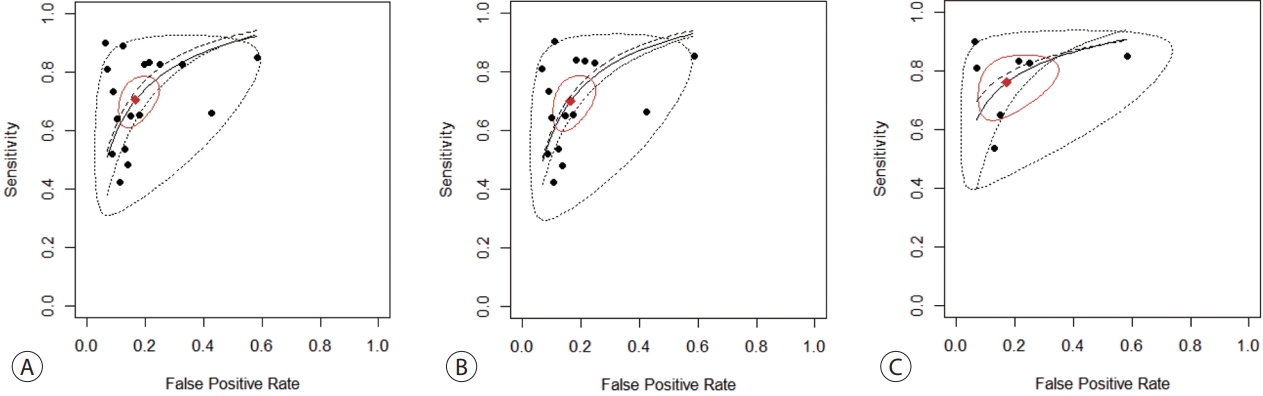
- The SENs and SPEs of various ctDNAs in the peripheral blood of patients with HBV-induced HCC were evaluated using forest plots (Fig. 3A). Significant heterogeneity was evident in terms of both SENs (I2=91.1%, P<0.001) and SPEs (I2=92.0%, P<0.001); the SENs and false-positive rates were positively correlated (rho 0.273; 95% confidence interval [CI], -0.239 to 0.667). The meta-analysis employed a random-effects model. The pooled sensitivity and specificity were 0.705 (95% CI, 0.629-0.771) and 0.833 (95% CI, 0.769-0.882), respectively. The DOR, PLR, and NLR were 11.759 (95% CI, 7.982-17.322), 4.285 (95% CI, 3.098-5.925), and 0.366 (95% CI, 0.301-0.336), respectively, and the SROC curve AUC was 0.835 (Fig. 4A).
- 4. Diagnostic value of ctDNA methylation assay in patients with HBV-induced HCC
- Of the nine studies, seven explored the diagnostic utility of ctDNA methylation assays. Significant heterogeneity was evident in terms of SENs (I2=89.7%, P<0.01) and SPEs (I2=89.1%, P<0.001). The pooled SEN and SPE were 0.7 (95% CI, 0.61-0.771) and 0.835 (95% CI, 0.768-0.885), respectively (Fig. 3B). The DOR, PLR, and NLR were 11.524 (95% CI, 7.539-17.616), 4.102 (95% CI, 2.925-5.752), and 0.379 (95% CI, 0.310-0.463), respectively. The SROC curve exhibited an AUC of 0.836 (Fig. 4B).
- 5. Diagnostic value of the ctDNA assay combined with tumor markers in patients with HBV-induced HCC
- Seven studies explored the diagnostic utility of combined ctDNA and tumor marker assays. One study measured both α-fetoprotein (AFP) and des-gamma-carboxy-prothrombin (DCP) levels, while the others considered only AFP levels. The I2 values were 66.9% for SENs and 85.0% for SPEs. The pooled SEN and SPE were 0.760 (95% CI, 0.659-0.839) and 0.828 (95% CI, 0.692-0.911), respectively (Fig. 3C), and the AUC of the SROC curve was 0.848 (Fig. 4C). The DOR, PLR, and NLR values were 14.273 (95% CI, 7.299-27.910), 4.641 (95% CI, 2.563-8.402), and 0.304 (95% CI, 0.215-0.430), respectively.
- 6. Meta-regression analysis and publication bias
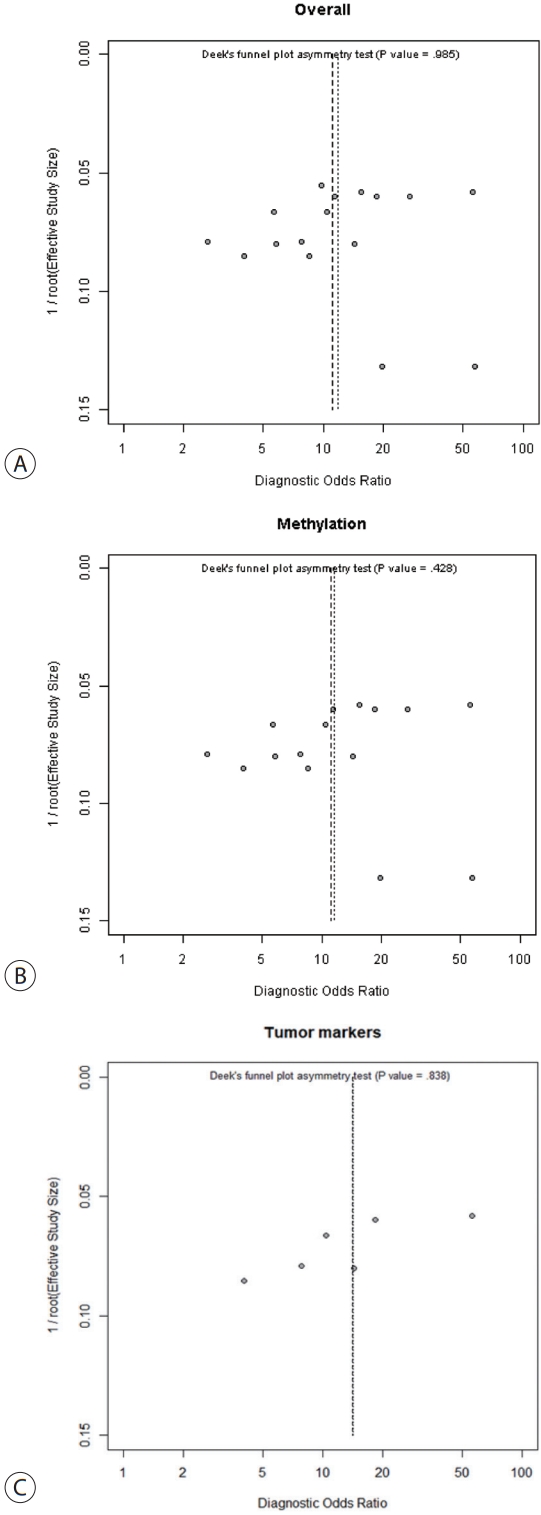
- Meta-regression analysis was performed to explore the potential causes of heterogeneity. Sample sources, assay methods, number of genes studied, and inclusion of tumor markers did not significantly contribute to heterogeneity (Table 2). Publication bias was assessed using the Deek funnel plot asymmetry test. No significant overall bias was evident (P=0.985) (Fig. 5A), and no bias was apparent in either the ctDNA methylation group (P=0.428) (Fig. 5B), or the ctDNA with tumor markers group (P=0.838) (Fig. 5C).
RESULTS
- HBV infection remains the principal risk factor for HCC and, thereby, for cancer-associated deaths. One of the factors primarily responsible for the poor prognosis of HCC is its low early diagnosis rate. Despite various surveillance programs, many patients are still diagnosed at advanced stages, when optimal treatment is often impossible. It is important to detect HCC at an early stage to increase the survival rate of patients.
- With the increasing advances in sequencing technology, novel tools for diagnosing liver diseases are emerging. Liquid biopsy is attracting attention as a noninvasive and reliable biomarker. Circulating extracellular vesicles, DNA, RNA, and tumor cells have emerged as attractive liquid biopsy candidates because they fulfil the key requirements of ideal biomarkers.39 The application of novel molecular techniques to liquid biopsies has improved our understanding of the impact of ctDNA detection on HCC diagnosis.40 We conducted a systematic meta-analysis of studies on ctDNA-based HCC diagnosis, particularly in patients with chronic hepatitis B.
- In this meta-analysis, various ctDNA assays, including somatic mutations and methylation, had a fair diagnostic performance for HBV-related HCC with an AUC of 0.835. The ctDNA methylation assay group showed a similar performance, with an AUC of 0.836. Notably, when ctDNA assays were combined with assays of tumor markers such as AFP and DCP, the diagnostic performance improved (AUCs of 0.838 and 0.829 with and without tumor marker assays, respectively; data not shown) in terms of discriminating HCC patients from control individuals. Previous studies have demonstrated that AFP exhibits unsatisfactory diagnostic performance (SENs, 0.478-0.540).41,42 When combined, detection of ctDNA and AFP assay showed an excellent diagnostic performance (AUC, 0.944).42 Along with our results, these results highlight that combinations of ctDNA and tumor markers have the potential to be novel adjuvant noninvasive tools for tumor markers in the screening and detection of HCC, regardless of its etiology.
- We measured the DORs, wherein a DOR >10 indicated that the test discrimination was satisfactory.43 The pooled DOR for the ctDNA assay that discriminated HCC patients from controls was 11.759, but this increased to 14.273 when the assay was combined with tumor markers, indicating the capability of integrating ctDNA assay with tumor markers to screen and detect HCC. The PLR and NLR values were also derived. The PLRs of the ctDNA assay alone and in combination with tumor marker assays were 4.285 and 4.641, respectively, indicating that HCC cases were 4- to 5-fold more likely to be ctDNA-positive than controls. The NLRs of the ctDNA assay alone and in combination with tumor marker assays were 0.366 and 0.304, respectively, indicating that the probability that individuals who tested negative in the ctDNA assay would in fact have HCC was 36.6% and 30.4%, respectively. Therefore, the negative ctDNA assay results should be interpreted with caution.
- This study had several limitations. First, despite a thorough literature search, several valuable articles might not have been included by the search strategy. Although we added these articles manually as much as possible, there is still the possibility that suitable publications were not included. Second, a significant among-study heterogeneity was observed. We performed a meta-regression to explore the potential causes of heterogeneity, but the sample source, assay method, number of genes studied, and selected tumor markers did not explain the heterogeneity. Heterogeneity might have arisen for other reasons that could not be assessed in this study because of a partial lack of data. Lastly, there are no data comparing the accuracy of ctDNA with surveillance versus surveillance alone for early HCC diagnosis. Further largescale prospective clinical studies are needed to confirm the diagnostic utility of ctDNA assays in HCC. In conclusion, ctDNA assays may aid in HCC screening and detection, particularly when combined with tumor marker assays.
DISCUSSION
-
Conflicts of Interest
Young Chang, Hyuksoo Eun and Do Seon Song currently serve as editorial board members for J Liver Cancer, and they were not involved in the review process of this article. Otherwise, the authors have no conflicts of interest to disclose.
-
Ethics Statement
This systemic review and meta-analysis is fully based on the articles which was already published and did not involve additional patient participants. Therefore, IRB approval is not necessary.
-
Funding Statement
This study was supported by the Soonchunhyang University Research Fund and the National Research Foundation of Korea grant funded by the Korea government (2020R1F1A1076282, 2021R1I1A3059993). This study was supported by the Research Award of the Korean Liver Cancer Association (2020).
-
Data Availability
The data presented in this study are available upon reasonable request from the corresponding author.
-
Author Contribution
Study concept and design: JSW, CY, PS
Acquisition of data: EH, SDS, LYS, CY, JSW
Analysis and interpretation of data: LHW, KSG, LSH, KW, YSJ, JJY, RS, CY, JSW
Drafting of the manuscript: CY, JSW
Statistical analysis: PS, CY, JSW
Study supervision: JSW, CY
Writing review & editing: all authors
Approval of final manuscript: all authors
Article information
Supplementary Material

| Study | Study type | Control type | HCC/LC/CHB/HC (number) | Sampling time | Sample source | Detection methods | Assay indicators | Cutoff value | SEN (%) | SPE (%) | TP | FP | FN | TN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cai et al. [31] (2019) | Case-control | CHB, LC | 220/129/0 | Pretreatment | Plasma | 5hmC-seal technique | wd-score | 27.9 | 82.7 | 67.4 | 182 | 42 | 38 | 87 |
| Qu et al. [19] (2019) | Case-control | CHB | 4/0/327/0 | Pretreatment | Plasma | NGS | TP53, CTNNB1, AXIN1, TERT, HBV integrations | NA | 100 | 94 | 4 | 20 | 0 | 307 |
| AFP, DCP | ||||||||||||||
| Dong et al. [32] (2017) | Case-control | CHB, LC, HC | 98/75/90/80 | NA | Serum | MSP | RASSF1A | NA | 52 | 91.5 | 51 | 21 | 47 | 224 |
| RASSF1A, BVES, HOXA9 | NA | 73.5 | 91.1 | 72 | 22 | 26 | 223 | |||||||
| RASSF1A, BVES, HOXA9, AFP | NA | 83.7 | 78.9 | 82 | 52 | 16 | 193 | |||||||
| Hu et al. [33] (2017) | Case-control | CHB, LC, HC | 80/40/40/0 | Pretreatment | Serum | MSP | UBE2Q1 | NA | 66.3 | 57.5 | 53 | 34 | 27 | 46 |
| UBE2Q1, AFP >200 ng/mL | NA | 53.8 | 87.5 | 43 | 10 | 37 | 70 | |||||||
| Dong et al. [34] (2015) | Case-control | CHB | 190/0/120/0 | Pretreatment | Serum | MSP | RASSF1A | NA | 64.2 | 89.8 | 122 | 12 | 68 | 108 |
| RASSF1A, AFP >20 ng/mL | NA | 80.9 | 93.4 | 154 | 8 | 36 | 112 | |||||||
| Li et al. [35] (2014) | Case-control | CHB | 136/0/46/0 | Pretreatment | Serum | MSP | IGFBP7 | NA | 65.4 | 82.6 | 89 | 8 | 47 | 38 |
| IGFBP7, AFP >20 ng/mL | NA | 85.3 | 41.3 | 116 | 27 | 20 | 19 | |||||||
| Han et al. [36] (2014) | Case-control | CHB | 160/0/88/0 | NA | Serum | MSP | TGR5 | NA | 48.1 | 86.4 | 77 | 12 | 83 | 76 |
| TGR5, AFP >400 ng/mL | NA | 65.0 | 85.2 | 104 | 13 | 56 | 75 | |||||||
| Yang et al. [37] (2014) | Case-control | CHB, LC | 123/28/29/0 | NA | Serum | MSP | CDO1 | NA | 42.3 | 89.5 | 52 | 6 | 71 | 51 |
| CDO1, AFP >20 ng/mL | NA | 82.9 | 75.4 | 102 | 14 | 21 | 43 | |||||||
| Zhang et al. [38] (2013) | Case-control | HC | 31/0/0/27 | Pretreatment | Serum | Illumina 450 beadchip and bisulfite sequencing | DBX2 | NA | 88.9 | 87.1 | 28 | 3 | 3 | 24 |
| THY1 | NA | 85.2 | 80.7 | 26 | 5 | 5 | 22 |
HCC, hepatocellular carcinoma; LC, liver cirrhosis; CHB, chronic hepatitis B; HC, healthy control; SEN, sensitivity; SPE, specificity; TP, true positive; FN, false negative; FP, false positive; TN, true negative; 5hmC, 5-hydroxymethylcytosines; NA, not available; NGS, next generation sequencing; MSP, methylation-specific polymerase chain reaction; wd score, weighted diagnostic score; TP53, tumor protein P53; CTNNB1, catenin beta 1; AXIN1, Axin 1; TERT, telomerase reverse transcriptase; HBV, hepatitis B virus; AFP, alpha-fetoprotein; DCP, des-gamma-carboxyprothrombin; RASSF1A, Ras association domain family member 1; BVES, blood vessel epicardial substance; HOXA9, homebox A9; UBE2Q1, ubiquitin conjugating enzyme E2 Q1; IGFBP7, insulin like growth factor binding protein 7; TGR5, G-protein-coupled bile acid receptor Gpbar1; CDO1, cysteine dioxygenase type 1; DBX2, developing brain homeobox 2; THY1, Thy-1 cell surface antigen.
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209−249.ArticlePubMedPDF
- 2. Center MM, Jemal A. International trends in liver cancer incidence rates. Cancer Epidemiol Biomarkers Prev 2011;20:2362−2368.ArticlePubMedPDF
- 3. Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: epidemiology, etiology, and carcinogenesis. J Carcinog 2017;16:1. ArticlePubMedPMC
- 4. Korean Liver Cancer Association, National Cancer Center. 2018 Korean Liver Cancer Association-National Cancer Center Korea practice guidelines for the management of hepatocellular carcinoma. Gut Liver 2019;13:227−299.ArticlePubMedPMC
- 5. European Association for the Study of the Liver. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182−236.ArticlePubMed
- 6. Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018;67:358−380.ArticlePubMedPDF
- 7. Chou R, Cuevas C, Fu R, Devine B, Wasson N, Ginsburg A, et al. Imaging techniques for the diagnosis of hepatocellular carcinoma: a systematic review and meta-analysis. Ann Intern Med 2015;162:697−711.ArticlePubMed
- 8. Singal AG, Nehra M, Adams-Huet B, Yopp AC, Tiro JA, Marrero JA, et al. Detection of hepatocellular carcinoma at advanced stages among patients in the HALT-C trial: where did surveillance fail? Am J Gastroenterol 2013;108:425−432.ArticlePubMedPMCPDF
- 9. Colli A, Fraquelli M, Casazza G, Massironi S, Colucci A, Conte D, et al. Accuracy of ultrasonography, spiral CT, magnetic resonance, and alpha-fetoprotein in diagnosing hepatocellular carcinoma: a systematic review. Am J Gastroenterol 2006;101:513−523.PubMed
- 10. Mezzalira S, De Mattia E, Guardascione M, Dalle Fratte C, Cecchin E, Toffoli G. Circulating-free DNA analysis in hepatocellular carcinoma: a promising strategy to improve patients’ management and therapy outcomes. Int J Mol Sci 2019;20:5498. ArticlePubMedPMC
- 11. Li X, Wang H, Li T, Wang L, Wu X, Liu J, et al. Circulating tumor DNA/circulating tumor cells and the applicability in different causes induced hepatocellular carcinoma. Curr Probl Cancer 2020;44:100516. ArticlePubMed
- 12. Alunni-Fabbroni M, Rönsch K, Huber T, Cyran CC, Seidensticker M, Mayerle J, et al. Circulating DNA as prognostic biomarker in patients with advanced hepatocellular carcinoma: a translational exploratory study from the SORAMIC trial. J Transl Med 2019;17:328. ArticlePubMedPMCPDF
- 13. Mody K, Kasi PM, Yang JD, Surapaneni PK, Ritter A, Roberts A, et al. Feasibility of circulating tumor DNA testing in hepatocellular carcinoma. J Gastrointest Oncol 2019;10:745−750.ArticlePubMedPMC
- 14. Chan KC, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013;59:211−224.ArticlePubMedPDF
- 15. Labgaa I, Villacorta-Martin C, D’Avola D, Craig AJ, von Felden J, Martins-Filho SN, et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene 2018;37:3740−3752.ArticlePubMedPMCPDF
- 16. Ng CKY, Di Costanzo GG, Tosti N, Paradiso V, Coto-Llerena M, Roscigno G, et al. Genetic profiling using plasma-derived cell-free DNA in therapy-naïve hepatocellular carcinoma patients: a pilot study. Ann Oncol 2018;29:1286−1291.ArticlePubMed
- 17. Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 2019;20:71−88.ArticlePubMedPDF
- 18. Cohen JD, Douville C, Dudley JC, Mog BJ, Popoli M, Ptak J, et al. Detection of low-frequency DNA variants by targeted sequencing of the Watson and Crick strands. Nat Biotechnol 2021;39:1220−1227.ArticlePubMedPMCPDF
- 19. Qu C, Wang Y, Wang P, Chen K, Wang M, Zeng H, et al. Detection of early-stage hepatocellular carcinoma in asymptomatic HBsAgseropositive individuals by liquid biopsy. Proc Natl Acad Sci U S A 2019;116:6308−6312.ArticlePubMedPMC
- 20. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926−930.ArticlePubMedPMC
- 21. Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV; CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol 2020;31:745−759.PubMed
- 22. Mah WC, Lee CG. DNA methylation: potential biomarker in hepatocellular carcinoma. Biomark Res 2014;2:5. ArticlePubMedPMCPDF
- 23. Adeniji N, Dhanasekaran R. Current and emerging tools for hepatocellular carcinoma surveillance. Hepatol Commun 2021;5:1972−1986.ArticlePubMedPMCPDF
- 24. Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet 2011;13:7−13.ArticlePubMedPDF
- 25. Zeng C, Stroup EK, Zhang Z, Chiu BC, Zhang W. Towards precision medicine: advances in 5-hydroxymethylcytosine cancer biomarker discovery in liquid biopsy. Cancer Commun (Lond) 2019;39:12. ArticlePubMedPMCPDF
- 26. Gao P, Lin S, Cai M, Zhu Y, Song Y, Sui Y, et al. 5-Hydroxymethylcytosine profiling from genomic and cell-free DNA for colorectal cancers patients. J Cell Mol Med 2019;23:3530−3537.ArticlePubMedPMCPDF
- 27. Li W, Zhang X, Lu X, You L, Song Y, Luo Z, et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res 2017;27:1243−1257.ArticlePubMedPMCPDF
- 28. Song CX, Yin S, Ma L, Wheeler A, Chen Y, Zhang Y, et al. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res 2017;27:1231−1242.ArticlePubMedPMCPDF
- 29. Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097.ArticlePubMedPMC
- 30. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529−536.ArticlePubMed
- 31. Cai J, Chen L, Zhang Z, Zhang X, Lu X, Liu W, et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut 2019;68:2195−2205.ArticlePubMedPMC
- 32. Dong X, Hou Q, Chen Y, Wang X. Diagnostic value of the methylation of multiple gene promoters in serum in hepatitis B virus-related hepatocellular carcinoma. Dis Markers 2017;2017:2929381. ArticlePubMedPMCPDF
- 33. Hu N, Fan XP, Fan YC, Chen LY, Qiao CY, Han LY, et al. Hypomethylated ubiquitin-conjugating enzyme2 Q1 (UBE2Q1) gene promoter in the serum is a promising biomarker for hepatitis B virus-associated hepatocellular carcinoma. Tohoku J Exp Med 2017;242:93−100.ArticlePubMed
- 34. Dong X, He H, Zhang W, Yu D, Wang X, Chen Y. Combination of serum RASSF1A methylation and AFP is a promising non-invasive biomarker for HCC patient with chronic HBV infection. Diagn Pathol 2015;10:133. ArticlePubMedPMCPDF
- 35. Li F, Fan YC, Gao S, Sun FK, Yang Y, Wang K. Methylation of serum insulin-like growth factor-binding protein 7 promoter in hepatitis B virus-associated hepatocellular carcinoma. Genes Chromosomes Cancer 2014;53:90−97.ArticlePubMed
- 36. Han LY, Fan YC, Mu NN, Gao S, Li F, Ji XF, et al. Aberrant DNA methylation of G-protein-coupled bile acid receptor Gpbar1 (TGR5) is a potential biomarker for hepatitis B Virus associated hepatocellular carcinoma. Int J Med Sci 2014;11:164−171.ArticlePubMedPMC
- 37. Yang Y, Fan YC, Gao S, Dou CY, Zhang JJ, Sun FK, et al. Methylated cysteine dioxygenase-1 gene promoter in the serum is a potential biomarker for hepatitis B virus-related hepatocellular carcinoma. Tohoku J Exp Med 2014;232:187−194.ArticlePubMed
- 38. Zhang P, Wen X, Gu F, Deng X, Li J, Dong J, et al. Methylation profiling of serum DNA from hepatocellular carcinoma patients using an Infinium Human Methylation 450 BeadChip. Hepatol Int 2013;7:893−900.ArticlePubMedPDF
- 39. Mann J, Reeves HL, Feldstein AE. Liquid biopsy for liver diseases. Gut 2018;67:2204−2212.ArticlePubMed
- 40. Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer 2019;18:114. ArticlePubMedPMCPDF
- 41. Farinati F, Marino D, De Giorgio M, Baldan A, Cantarini M, Cursaro C, et al. Diagnostic and prognostic role of alpha-fetoprotein in hepatocellular carcinoma: both or neither? Am J Gastroenterol 2006;101:524−532.ArticlePubMed
- 42. Zhang Z, Chen P, Xie H, Cao P. Using circulating tumor DNA as a novel biomarker to screen and diagnose hepatocellular carcinoma: a systematic review and meta-analysis. Cancer Med 2020;9:1349−1364.ArticlePubMedPMCPDF
- 43. Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol 2003;56:1129−1135.ArticlePubMed
References
Figure & Data
References
Citations

- 16S rRNA Next-Generation Sequencing May Not Be Useful for Examining Suspected Cases of Spontaneous Bacterial Peritonitis
Chan Jin Yang, Ju Sun Song, Jeong-Ju Yoo, Keun Woo Park, Jina Yun, Sang Gyune Kim, Young Seok Kim
Medicina.2024; 60(2): 289. CrossRef - Methylated circulating tumor DNA in hepatocellular carcinoma: A comprehensive analysis of biomarker potential and clinical implications
Qian Zhu, Jiaqi Xie, Wuxuan Mei, Changchun Zeng
Cancer Treatment Reviews.2024; 128: 102763. CrossRef

 E-submission
E-submission THE KOREAN LIVER CANCER ASSOCIATION
THE KOREAN LIVER CANCER ASSOCIATION
 PubReader
PubReader ePub Link
ePub Link Download Citation
Download Citation

 Follow JLC on Twitter
Follow JLC on Twitter