Search
- Page Path
- HOME > Search
Original Articles
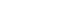
- The diagnostic value of circulating tumor DNA in hepatitis B virus induced hepatocellular carcinoma: a systematic review and meta-analysis
- Young Chang, Soung Won Jeong, Jae Young Jang, Hyuksoo Eun, Young‑Sun Lee, Do Seon Song, Su Jong Yu, Sae Hwan Lee, Won Kim, Hyun Woong Lee, Sang Gyune Kim, Seongho Ryu, Suyeon Park
- J Liver Cancer. 2022;22(2):167-177. Published online September 29, 2022
- DOI: https://doi.org/10.17998/jlc.2022.09.19
- 2,649 Views
- 74 Downloads
- 1 Citation
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background/Aim
New biomarkers are urgently needed to aid in the diagnosis of early stage hepatocellular carcinoma (HCC). We performed a meta-analysis on the diagnostic utility of circulating tumor DNA (ctDNA) levels in patients with hepatitis B virus-induced HCC.
Methods
We retrieved relevant articles from PubMed, Embase, and the Cochrane Library up to February 8, 2022. Two subgroups were defined; one subset of studies analyzed the ctDNA methylation status, and the other subset combined tumor markers and ctDNA assays. Pooled sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under the summary receiver operating characteristic curve (AUC) were analyzed.
Results
Nine articles including 2,161 participants were included. The overall SEN and SPE were 0.705 (95% confidence interval [CI], 0.629-0.771) and 0.833 (95% CI, 0.769-0.882), respectively. The DOR, PLR, and NLR were 11.759 (95% CI, 7.982-17.322), 4.285 (95% CI, 3.098- 5.925), and 0.336 (0.301-0.366), respectively. The ctDNA assay subset exhibited an AUC of 0.835. The AUC of the combined tumor marker and ctDNA assay was 0.848, with an SEN of 0.761 (95% CI, 0.659-0.839) and an SPE of 0.828 (95% CI, 0.692-0.911).
Conclusions
Circulating tumor DNA has promising diagnostic potential for HCC. It can serve as an auxiliary tool for HCC screening and detection, especially when combined with tumor markers. -
Citations
Citations to this article as recorded by- 16S rRNA Next-Generation Sequencing May Not Be Useful for Examining Suspected Cases of Spontaneous Bacterial Peritonitis
Chan Jin Yang, Ju Sun Song, Jeong-Ju Yoo, Keun Woo Park, Jina Yun, Sang Gyune Kim, Young Seok Kim
Medicina.2024; 60(2): 289. CrossRef
- 16S rRNA Next-Generation Sequencing May Not Be Useful for Examining Suspected Cases of Spontaneous Bacterial Peritonitis

- Diagnostic performance of serum exosomal miRNA-720 in hepatocellular carcinoma
- Jeong Won Jang, Ji Min Kim, Hye Seon Kim, Jin Seoub Kim, Ji Won Han, Soon Kyu Lee, Heechul Nam, Pil Soo Sung, Si Hyun Bae, Jong Young Choi, Seung Kew Yoon
- J Liver Cancer. 2022;22(1):30-39. Published online March 21, 2022
- DOI: https://doi.org/10.17998/jlc.2022.02.25
- 3,938 Views
- 136 Downloads
- 1 Citation
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background/Aim
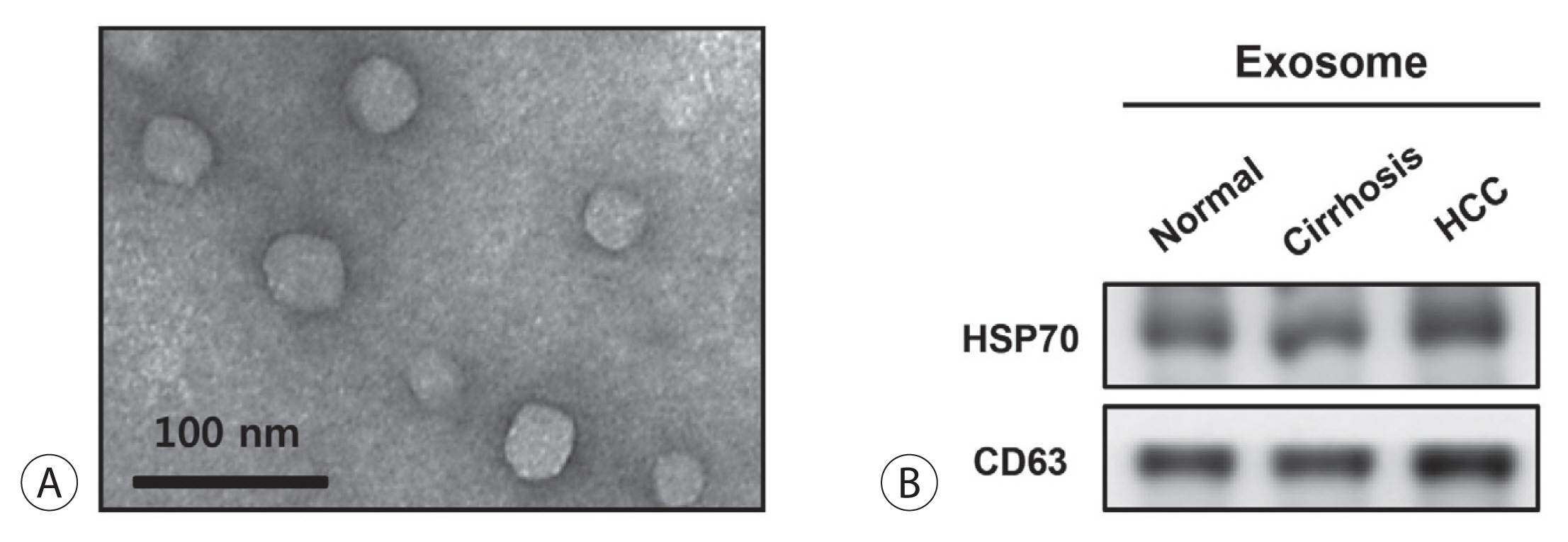
Hepatocellular carcinoma (HCC) is associated with poor prognosis, largely due to late detection. Highly accurate biomarkers are urgently needed to detect early-stage HCC. Our study aims to explore the diagnostic performance of serum exosomal microRNA (miR)-720 in HCC.
Methods
Exosomal miRNA was measured via quantitative real-time PCR. A correlation analysis of exosomal miR-720 and tumor or clinico-demographic data of patients with HCC was performed. The receiver operating characteristic (ROC) curve was used to assess the diagnostic capacity of serum exosomal miR-720 for HCC, in comparison with α-fetoprotein (AFP) and prothrombin induced by vitamin K absence or antagonist-II (PIVKA-II).
Results
MiR-720 was chosen as a potential HCC marker via miR microarray based on significant differential expression between tumor and non-tumor samples. Serum exosomal miR-720 was significantly upregulated in patients with HCC (n=114) versus other liver diseases (control, n=30), with a higher area under the ROC curve (AUC=0.931) than the other markers. Particularly, serum exosomal miR-720 showed superior performance in diagnosing small HCC (< 5 cm; AUC=0.930) compared with AFP (AUC=0.802) or PIVKA-II (AUC=0.718). Exosomal miR-720 levels showed marginal correlation with tumor size. The proportion of elevated miR-720 also increased with intrahepatic tumor stage progression. Unlike AFP or PIVKA-II showing a significant correlation with aminotransferase levels, the exosomal miR-720 level was not affected by aminotransferase levels.
Conclusions
Serum exosomal miR-720 is an excellent biomarker for the diagnosis of HCC, with better performance than AFP or PIVKA-II. Its diagnostic utility is maintained even in small HCC and is unaffected by aminotransferase levels. -
Citations
Citations to this article as recorded by- Prospects of liquid biopsy in the prognosis and clinical management of gastrointestinal cancers
Deepankar Mondal, Sapnita Shinde, Vibha Sinha, Vineeta Dixit, Souvik Paul, Rakesh Kumar Gupta, Suresh Thakur, Naveen Kumar Vishvakarma, Dhananjay Shukla
Frontiers in Molecular Biosciences.2024;[Epub] CrossRef
- Prospects of liquid biopsy in the prognosis and clinical management of gastrointestinal cancers

- Gut-microbiome Taxonomic Profiling as Non-invasive Biomarkers for the Early Detection of Alcoholic Hepatocellular Carcinoma
- Jun Seok, Ki Tae Suk
- J Liver Cancer. 2020;20(1):32-40. Published online March 31, 2020
- DOI: https://doi.org/10.17998/jlc.20.1.32

- 5,473 Views
- 235 Downloads
-
 Abstract
Abstract
 PDF
PDF - Background/Aims
Hepatocellular carcinoma (HCC) is a prevalent form of primary liver cancer and the fifth leading cause of worldwide cancer mortality. Though early diagnosis of HCC is important, so far lack of effective biomarkers for early diagnosis of HCC has been a problem. In this study, we searched for potential functional biomarkers of alcoholic HCC by using metagenomics approach.
Methods
Between September 2017 and April 2019, normal control (n=44), alcoholic liver cirrhosis (n=44), and alcoholic HCC (n=13) groups were prospectively enrolled and analyzed. Gut microbiota was analyzed using the 16S-based microbiome taxonomic profiling platform of EzBioCloud Apps and analyzing system.
Results
There was a statistically significant difference among groups in diversity (P<0.05). In the comparison of phylum between cirrhosis and HCC, Proteobacteria were increased and Bacteroidetes were decreased. Firmicutes were not significantly different among the three groups. In the taxonomic profiling, relative abundance of Lactobacillus in the cirrhosis and HCC groups showed richness (P<0.05). In the biomarker analysis between cirrhosis and HCC, obiquinome Fe-S protein 3, global nitrogen regulator, Vesicle-associated membrane protein 7, toxin YoeB, peroxisome-assembly ATPase, and nitrogen oxide reductase regulator were differently expressed (P<0.001).
Conclusions
Alcoholic HCC showed different expressions in the stool taxonomy and biomarker compared with that of cirrhosis and control. Therefore, new biomarkers using stool analysis for alcoholic HCC are necessary.


 E-submission
E-submission THE KOREAN LIVER CANCER ASSOCIATION
THE KOREAN LIVER CANCER ASSOCIATION

 First
First Prev
Prev



 Follow JLC on Twitter
Follow JLC on Twitter