Articles
- Page Path
- HOME > J Liver Cancer > Volume 22(1); 2022 > Article
-
Original Article
Diagnostic performance of serum exosomal miRNA-720 in hepatocellular carcinoma -
Jeong Won Jang1,2
 , Ji Min Kim2,3
, Ji Min Kim2,3 , Hye Seon Kim2,3
, Hye Seon Kim2,3 , Jin Seoub Kim2,3
, Jin Seoub Kim2,3 , Ji Won Han1,2
, Ji Won Han1,2 , Soon Kyu Lee1,2
, Soon Kyu Lee1,2 , Heechul Nam1,2
, Heechul Nam1,2 , Pil Soo Sung1,2
, Pil Soo Sung1,2 , Si Hyun Bae1,2
, Si Hyun Bae1,2 , Jong Young Choi1,2
, Jong Young Choi1,2 , Seung Kew Yoon1,2
, Seung Kew Yoon1,2
-
Journal of Liver Cancer 2022;22(1):30-39.
DOI: https://doi.org/10.17998/jlc.2022.02.25
Published online: March 21, 2022
1Department of Internal Medicine, The Catholic University of Korea, Seoul, Korea
2The Catholic University Liver Research Center, The Catholic University of Korea, Seoul, Korea
3Department of Biomedicine & Health Sciences, College of Medicine, The Catholic University of Korea, Seoul, Korea
-
Corresponding author: Jeong Won Jang, Division of Hepatology, Department of Internal Medicine, Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, 222 Banpo-daero, Seocho-gu, Seoul 06591, Korea,
Tel. +82-2-2258-6015, Fax. +82-2-3481-4025, E-mail: garden@catholic.ac.kr
Copyright © 2022 by The Korean Liver Cancer Association
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- 3,758 Views
- 131 Downloads
Abstract
-
Background/Aim
- Hepatocellular carcinoma (HCC) is associated with poor prognosis, largely due to late detection. Highly accurate biomarkers are urgently needed to detect early-stage HCC. Our study aims to explore the diagnostic performance of serum exosomal microRNA (miR)-720 in HCC.
-
Methods
- Exosomal miRNA was measured via quantitative real-time PCR. A correlation analysis of exosomal miR-720 and tumor or clinico-demographic data of patients with HCC was performed. The receiver operating characteristic (ROC) curve was used to assess the diagnostic capacity of serum exosomal miR-720 for HCC, in comparison with α-fetoprotein (AFP) and prothrombin induced by vitamin K absence or antagonist-II (PIVKA-II).
-
Results
- MiR-720 was chosen as a potential HCC marker via miR microarray based on significant differential expression between tumor and non-tumor samples. Serum exosomal miR-720 was significantly upregulated in patients with HCC (n=114) versus other liver diseases (control, n=30), with a higher area under the ROC curve (AUC, 0.931) than the other markers. Particularly, serum exosomal miR-720 showed superior performance in diagnosing small HCC (<5 cm; AUC, 0.930) compared with AFP (AUC, 0.802) or PIVKA-II (AUC, 0.718). Exosomal miR-720 levels showed marginal correlation with tumor size. The proportion of elevated miR-720 also increased with intrahepatic tumor stage progression. Unlike AFP or PIVKA-II showing a significant correlation with aminotransferase levels, the exosomal miR-720 level was not affected by aminotransferase levels.
-
Conclusions
- Serum exosomal miR-720 is an excellent biomarker for the diagnosis of HCC, with better performance than AFP or PIVKA-II. Its diagnostic utility is maintained even in small HCC and is unaffected by aminotransferase levels.
- Hepatocellular carcinoma (HCC) is one of the most common primary liver cancers worldwide. Despite advances in the treatment of HCC, the prognosis of HCC still remains dismal, largely due to delayed detection. A number of biomarkers have been proposed to establish HCC diagnosis. Among these, α-fetoprotein (AFP) and prothrombin-induced by vitamin K absence or antagonist-II (PIVKA-II) are the most frequently used markers for HCC, but have several limitations, including false positivity, low sensitivity, and especially low detectability in early-stage HCC.1,2 Thus, there is the need for highly accurate biomarkers for HCC to enable prompt and accurate diagnosis, and subsequent improvement in overall survival outcomes.
- MicroRNAs (miRNAs) are endogenous small, single-stranded non-coding RNA molecules that exert RNA silencing and regulatory effects on gene expression through base pairing with complementary sequences within mRNA molecules. Studies have shown that miRNAs are involved in various biological processes, including hepatocarcinogenesis.3,4 They play a key regulatory role in cell proliferation, apoptosis, invasion, metastasis, epithelial-mesenchymal transition, angiogenesis, drug resistance and autophagy in HCC.3
- Although a number of studies have compiled miRNAs for measurement of varying outcomes in HCC,3 fewer studies involved exosomal miRNAs. Exosomes are 30- to 150-nm-sized extracellular vesicles that are generated in the endosomal compartment of cells. They contain unique protein and RNA cargo, and are secreted from a wide variety of cells into biological fluids, including blood, urine and cerebrospinal fluid.5,6 Unlike conventional circulating miRNAs, exosome-encapsulated miRNAs have several advantages as biomarkers in that they are resistant to RNases, and thus represent a highly stable and a rich source of biomarkers in biofluids.6 Recently, they have been suggested as better biomarkers for HCC than their serum-free counterparts.7 Additionally, given the similarities in miRNA content of exosomes and originating cancer cells,8 exosomal miRNA testing is highly specific towards the tumor environment than cell-free miRNAs directly measured in serum or plasma. Furthermore, cancer cells secrete a substantially higher number of exosomes than do normal cells, facilitating the transfer of oncogenic signals and tumorigenesis via cell-to-cell communication. 9 Taken together, exosomal miRNAs represent a promising biomarker in cancer for further investigation.
- Thus, this study aimed to investigate the role of circulating exosomal miRNAs in the diagnosis of HCC. We performed miRNA array and identified a candidate serum exosomal miRNA as a biomarker for early detection of HCC. The diagnostic utility of the exosomal miRNA was tested in comparison with AFP and PIVKA-II and correlation with HCC characteristics.
INTRODUCTION
- 1. Patients and samples
- The current study recruited a total of 144 subjects, including 114 patients with HCC and 30 without HCC. All of the HCC patients had confirmed HCC diagnosis based on typical imaging findings or pathological examination of tumors.10 Tumor stage was assessed according to the modified Union for International Cancer Control (mUICC) stage, which was adopted in the Korean National Cancer Center practice guideline.10 Our study also included serum samples from 30 non-HCC patients as a control group. Each patient provided informed consent to participate in the study. This study was approved by the Ethics Committees of The Catholic University of Korea (KC16TISI0041) and written informed consent was obtained from all the participants. The Strengthening the Reporting of Observational studies in Epidemiology (STROBE) reporting guidelines were followed (Supplementary Table 1).
- 2. miRNA microarray
- For the miRNA microarray, we collected blood samples from four patients with HCC and four non-HCC patients. The total RNA was extracted from serum and the quality control of RNA samples was performed to assess quantity, quality, and purity. Each sample was labeled with alkaline phosphatase and hybridized using an Agilent hybridization system with Agilent Mouse miRNA v17.0 array. The miRNA expression profiling was analyzed with GeneSpring GX v11.5.1 (Agilent Technologies, Santa Clara, CA, USA).
- 3. Blood samples and exosome isolation
- Blood samples were routinely collected from the study subjects at the diagnosis of liver disease and were stored at −80°C. Exosome isolation from sera was conducted using ExoQuick™ (System Biosciences, Palo Alto, CA, USA). In brief, the frozen sera were thawed and then centrifuged at 3,000 g for 15 minutes at 4°C to remove cellular debris. Exosomes were then isolated from sera according to the manufacturer’s instructions. Exosome characterization was performed by resuspending the exosome pellet in PBS and aliquoted for visualization with transmission electron microscopy (TEM). Further characterization of exosomes by size and physical property as well as exosomal markers was described previously.11
- 4. Quantitative analysis of exosomal miRNA
- For quantitative analysis of exosomal miRNA, cDNA synthesis was performed with TaqMan™ microRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). The expression of miRNAs was analyzed with quantitative real-time PCR (qPCR) using TaqMan™ Universal PCR Master Mix, No AmpErase™ UNG (Applied Biosystems) according to the manufacturer’s instructions. The qPCR conditions were as follows: 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. The data were normalized to hsa-miR-16 and calculated via 2−ΔΔCT method. Each sample was analyzed in triplicate.
- 5. Statistical analysis
- Data were presented as mean±standard deviation or median (interquartile range, IQR). Comparisons between groups were appropriately performed using Student’s t-test, Mann-Whitney U test, ANOVA test, Kruskal-Wallis test, or chi-square test, when appropriate. Continuous variables were dichotomized based on their median values. The optimal cut-off level of exosomal miRNA for diagnosis of HCC was determined using the Youden index. The sensitivity and specificity of the exosomal miRNA were calculated using standard formulae. Receiver operating characteristic (ROC) curves and area under the ROC curve (AUC) were established to distinguish patients with HCC from those without HCC. A P-value <0.05 was considered significant. Statistical analysis and graphic design were conducted using IBM SPSS version 24.0.0.0 (IBM, Armonk, NY, USA) and PRISM GraphPad version 5.00 (GraphPad Software, San Diego, CA, USA).
METHOD
- 1. Patient characteristics
- Patients with HCC included 85 males (74.6%) and 29 females (25.4%), and aged 59.7±11.6 years, while the control group consisted of 17 males (56.7%) and 13 females (43.3%), and aged 51.1±15.6 years. For the HCC group, the mean tumor size was 7.7±5.6 cm and 62 patients (54.4%) had multiple tumors. Tumor stage in the 114 HCC patients included I in 13 (11.4%), II in 24 (21.1%), III in 28 (24.6%), IVa in 17 (14.9%), and IVb in 32 patients (28.1%). The majority of patients had HBV-associated HCC (n=88; 77.2%) and Child-Turcotte-Pugh (CTP) class A function (n=87; 76.3%). The baseline characteristics of the 144 study subjects (HCC: n=114 and non-HCC controls: n=30) are shown in Table 1.
- 2. Identification and characterization of exosomes
- Using TEM, we purified round vesicles, each measuring 30–150 nm in diameter, which suggested that the isolated vesicles were likely exosomes (Fig. 1A). In addition, western blot analysis showed enrichment of exosomal markers including CD63 (Santa Cruz Biotechnology, CA, USA, sc15363) and HSP70 (Enzo life sciences, Farmingdale, NY, USA; C92F3A-5) (Fig. 1B). These results demonstrated that the particles isolated from the sera of the study subjects were exosomes.
- 3. miRNA profiling and identification of serum exosomal miRNA-720 (miR-720)
- We performed miRNA microarray analyses of samples from four patients with HCC and from four patients with non-HCC chronic liver disease, as previously described.11 Ten miRNAs including miR-720, miR-1274-b, miR-4286, miR-1260, miR-1260b, miR-1274a, miR-25, miR-320a, miR-501-5p, and miR-130a were differentially expressed between HCC and non-HCC samples. Among the miRNAs, miR-720 that was significantly upregulated (fold change >1.5 and P-value <0.05) in HCC patients compared with non-HCC patients was finally selected for further analyses.
- 4. Serum exosomal miR-720 levels in HCC and non-HCC groups
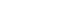
- Overall, the serum levels of exosomal miR-720 were significantly higher in patients with HCC than those without HCC (P <0.0001; Fig. 2A). Based on the analysis of ROC curve, exosomal miR-720 levels differed clearly between patients with and without HCC, with the AUC of 0.931 (95% confidence interval [CI], 0.881–0.981; P <0.001), which was better than that of AFP (AUC, 0.857) or PIVKA-II (AUC, 0.848) (Fig. 2B). The sensitivity and specificity of serum exosomal miR-720 were 86.0% and 82.4%, respectively, for discriminating HCC from non-HCC controls, when the cut-off point was set at 2.52 (Table 2). Consistently, exosomal miR-720 showed better performance in discriminating between HCC and non-HCC liver diseases compared to AFP or PIVKA-II within patients with chronic HBV infection (Supplementary Fig. 1).
- 5. Discriminative ability of serum exosomal miR-720 for small HCC
- We then investigated whether serum exosomal miR-720 discriminated small HCC (<5 cm) from larger ones successfully. The results showed that serum exosomal miR-720 had better discriminative ability with an AUC of 0.930 (95% CI, 0.868–0.992; P <0.001) between small HCCs (<5 cm) and non-HCC liver diseases than AFP (AUC, 0.802) or PIVKA-II (AUC, 0.718) (Fig. 2C). When the cut-off was set at 4.30, the sensitivity was 80.4% and the specificity was 88.2% in discriminating small HCC from non-tumor liver diseases (Table 2). We also found better discriminative ability of exo-somal miR-720 than AFP or PIVKA-II for HCC smaller than 2 cm (Supplementary Fig. 2). The overall findings suggest the excellent performance of circulating exosomal miR-720 as a diagnostic biomarker in discriminating HCC patients from those with chronic liver disease.
- 6. Association of serum exosomal miR-720 with HCC characteristics
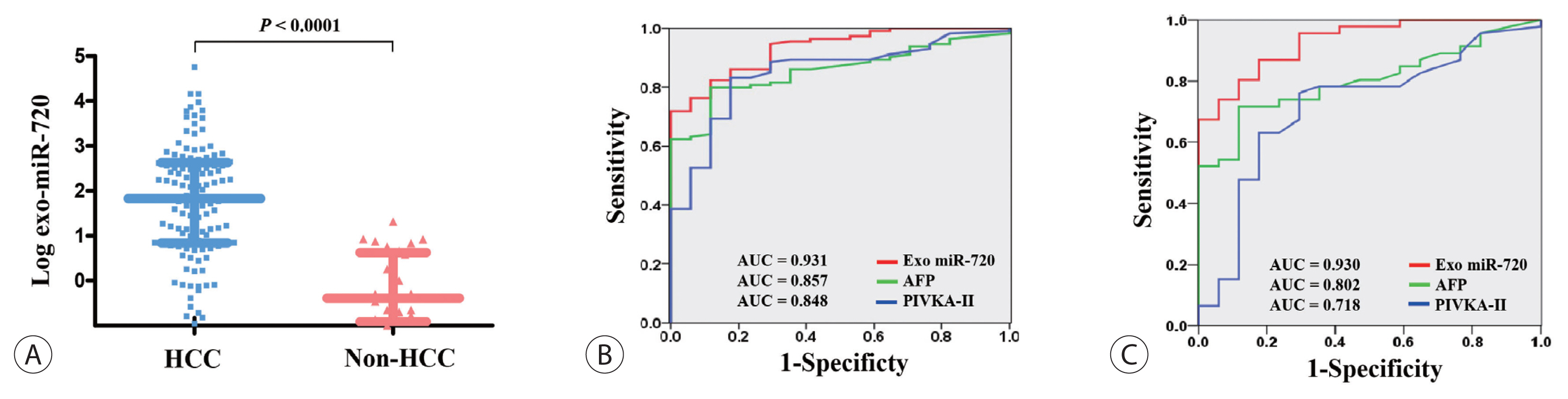
- The association between serum exosomal miR-720 levels and tumor characteristics was analyzed and the results are shown in Fig. 3. When plotted according to tumor size, a positive correlation was observed between the levels of exosomal miR-720 and the size of HCC, but it did not reach statistical significance (P =0.0629 and r=0.2103; Fig. 3A). The levels of serum exosomal miR-720 were not significantly different between patients with a single tumor and those with multiple tumors (1.53 and 2.20, respectively; P =0.2564; Fig. 3B). However, the exosomal miR-720 levels tended to increase with intrahepatic tumor stage, from mUICC stage I to IVa (P =0.3006; Fig. 3C), Although no definitive linear correlation with mUICC HCC stage from I to IV was observed, patients with stages I or II HCC carried lower serum exosomal miR-720 levels than those with stages III or IV (1.18 and 2.18, respectively; P =0.1361). The proportion of patients with high exosomal miR-720 (>52.4, the median) was significantly increased with more advanced stage (P for trend= 0.017; Fig. 3D). Our data revealed no association between exosomal miR-720 levels and other tumor characteristics, such as portal vein invasion or extrahepatic metastasis (data not shown).
- 7. Association of serum exosomal miR-720 with demographic features
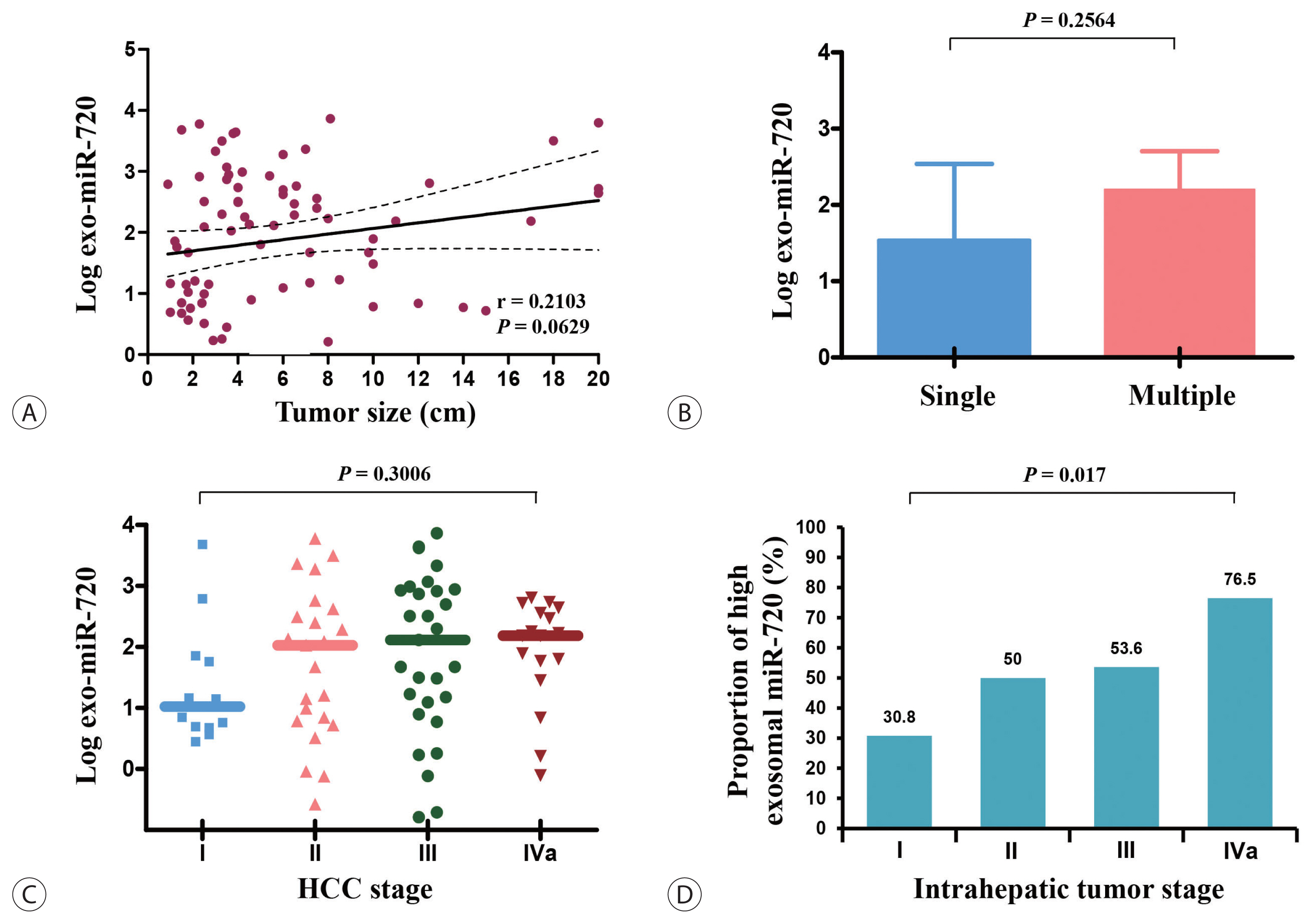
- Lastly, we analyzed the differences in exosomal miR-720 levels according to demographic data of HCC patients. The expression levels of circulating exosomal miR-720 were significantly higher in younger patients than in older patients (P =0.0064; Fig. 4A) as well as in patients with HBV-associated HCC than in those with non-HBV HCC (P =0.0488; Fig. 4B). The levels of exosomal miR-720 did not differ across the CTP classes (Fig. 4C), another important prognostic factor for HCC. Regarding liver inflammation, no significant correlations were found between serum exosomal miR-720 levels and aspartate aminotransferase (AST) or alanine aminotransferase (ALT) levels (P >0.05), whereas serum AFP or PIVKA-II levels exhibited significant positive linear correlation with AST or ALT levels (all P <0.05; Fig. 4D, E), indicating their limited diagnostic ability for HCC in patients with high aminotransferase levels.
RESULTS
- This study explored the role of a novel serum marker, exosomal miR-720, in HCC diagnosis. Using microarray analysis of miRNAs, miR-720 was selected as a potential marker due to its significantly differential expression among the HCC and non-HCC samples. The study results showed a significantly elevated expression of circulating exosomal miR-720 in HCC versus non-HCC subjects. The ROC curves of exosomal miR-720 used to distinguish patients with HCC from those without HCC were superior to AFP and PIVKA-II. In particular, the diagnostic potential of exosomal miR-720 was excellent even for small HCC in contrast to AFP or PIVKA-II, which showed only suboptimal efficiency to detect such cases. In contrast to the other markers, exosomal miR-720 was not affected by AST or ALT levels, and in general correlated with tumor characteristics. The overall findings indicate the excellent performance of circulating exosomal miR-720 as a diagnostic marker for HCC.
- AFP is currently the most widely used biomarker for HCC. However, it has limitations as a marker for screening HCC, due to its poor sensitivity (40–60%) and specificity (80–94%), when using the cut-off of 20 ng/mL12,13 and approximately 30–50% of the patients have normal AFP levels, especially in those with early-stage or small HCCs.1,13,14 PIVKA-II also remains unsatisfactory as a tumor marker for HCC due to its elevated concentration in patients with obstructive jaundice, alcohol intake, vitamin K deficiency, or warfarin therapy.2,15 In this regard, it is remarkable that circulating levels of exosomal miR-720 showed excellent diagnostic performance with a better ROC curve than AFP or PIVKA-II (Fig. 2). Moreover, the results appear relevant since the control group in this study included patients with chronic liver disease under actual HCC surveillance. The ROC of exosomal miR-720 was excellent, exceeding 0.9, which represents one of the best diagnostic performances as a marker for HCC ever reported.
- The diagnosis of early HCC is clinically of paramount importance as it contributes to treatments with curative intent and excellent long-term prognosis for patients with HCC.10 Our results showed that exosomal miR-720 had significant comparative advantages over AFP or PIVKA-II in detecting small HCC (<5 cm). Importantly, serum exosomal miR-720 exhibited a still high AUC (0.930) when used for distinguishing small HCC, whereas AFP or PIVKA-II yielded significantly decreased AUCs for such cases compared with those for overall HCC cases. Thus, it would be interesting to investigate the role of exosomal miR-720 as an adjunct screening tool, especially for small HCC.
- Based on our results, the excellent diagnostic performance of exosomal miR-720 in HCC is attributed to the levels of exosomal miR-720, which are not affected by liver inflammation. Diagnostic accuracy of other tumor markers such as AFP or PIVKA-II for HCC is negated in patients with liver inflammation, due to its false positivity in patients with elevated AST or ALT levels. Indeed, our results showed that AFP or PIVKA-II levels had significant positive correlations with both AST and ALT (Fig. 4D, E), yielding false positive results. However, unlike AFP or PIVKA-II, the exosomal miR-720 levels were not affected by aminotransferase concentrations, suggesting that exosomal miR-720 testing rather than AFP or PIVKA-II can be recommended for HCC screening in patients with elevated aminotransferase levels.
- In addition to the diagnostic role discussed above, it will be interesting to investigate whether exosomal miR-720 plays a role in treatment response and guidance for HCC treatment allocation. The potential function of exosomal miR-720 as a biomarker is supported by our findings correlating exosomal miR-720 with the size and progression of intrahepatic HCC stage. Additionally, our analysis is interesting in that the level of exosomal miR-720 was significantly higher in younger patients and in those with HBV-associated HCC. Future studies are required to determine whether the performance of exosomal miR-720 varies with age and the etiology of liver disease.
- miR-720 is located on chromosome 3q26.1 and is reportedly not a classic miRNA, but probably a tRNA fragment.16 Earlier studies reported conflicting results of oncogenic function of miR-720. High miR-720 expression was reported to positively correlate with higher pathologic stage and poor overall survival of patients with renal cell carcinoma17 or colorectal cancer18 as well as promote the aggressive phenotype of triple-negative breast cancer cells.19 Increasing levels of miR-720 was associated with glioma migration and invasion through downregulation of TARSL2,20 predicting adverse prognosis.21 In contrast, for breast cancer, miR-720 reportedly inhibited invasion and migration via directly targeting TWIST1,22 suggesting the complicated functions of miR-720 in tumorigenesis. To date, very few studies of exosomal miR-720 are available and no study has so far explored the role of exosomal miR-720 in the context of HCC. Given the relationship of exosomal miR-720 with HCC tumor characteristics, the biological role and function of miR-720 in hepatocarcinogenesis deserve further investigation.
- Our study has several limitations. The retrospective nature of this study is associated with an inherent selection bias. Thus, the study subjects are heterogeneous with a considerable proportion of patients in advanced stages of HCC. The sample size of the study is relatively small. Lastly, it is a cross-sectional study, lacking serial data related to exosomal miR-720 levels, and thus the changing levels during HCC surveillance or treatment should be evaluated to confirm exosomal miR-720 as a convincing marker of HCC. Nevertheless, the strength of our study is inclusion of subjects with chronic liver disease, but not healthy volunteers, as a control group, which may enhance its diagnostic reliability in clinical practice. More importantly, we identified a novel exosome-based biomarker, miR-720, based on a panel of miRNAs, reported here for the first time in the setting of HCC.
- In conclusion, the present study demonstrates the strong diagnostic performance of exosomal miR-720 in HCC. In particular, exosomal miR-720 can be effectively used to diagnose patients with small HCC or elevated aminotransferase levels. It remains an open question whether exosomal miR-720 testing combined with other markers may improve the diagnostic accuracy of HCC and represents a surrogate indicator of HCC prognosis. To address these issues, the biological properties of exosomal miR-720 should be further investigated in future functional studies.
DISCUSSION
-
Conflicts of Interest
The authors have no financial or personal relationships with other persons or organizations that could inappropriately affect the work. Jeong Won Jang has served as a consultant to or has served on the advisory board of Gilead, BMS, Abbvie, Bayer, Ipsen, and Roche.
-
Ethics Statement
This study was approved by the Ethics Committees of The Catholic University of Korea (KC16TISI0041) and written informed consent was obtained from all the participants.
-
Funding Statement
This study was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2021R1I1A2056660). This study was supported by the Research Award of the Korean Liver Cancer Association (2020).
-
Author Contribution
Study concept and design: JWJ
Acquisition of data: JWH, SKL, HN, PSS, SHB, JYC, SKY
Analysis and interpretation of data: JWJ, JMK, HSK, JSK
Drafting of the manuscript: JWJ
Statistical analysis: JWJ, JMK, HSK, JSK
Study supervision: JWJ
Article information
Supplementary Material

| HCC group (n=114) | Control group (n=30) | |
|---|---|---|
| Sex | ||
| Male | 85 (74.6) | 17 (56.7) |
| Female | 29 (25.4) | 13 (43.3) |
|
|
||
| Age (years) | 58.1±12.2 | 52.2±12.6 |
|
|
||
| Cause of liver disease | ||
| HBV | 87 (76.3) | 8 (26.7) |
| HCV | 12 (10.5) | 0 (0.0) |
| Non-viral | 15 (13.2) | 22 (73.3)* |
|
|
||
| AST (IU/L) | 57 (33–95) | 50 (28–83) |
|
|
||
| ALT (IU/L) | 38 (25–65) | 45 (20–170) |
|
|
||
| Child-Pugh class | ||
| A | 87 (76.3) | |
| B/C | 27 (23.7) | |
|
|
||
| Tumor size (cm) | 7.7±5.6 | |
|
|
||
| Tumor number | ||
| Single | 52 (45.6) | |
| Multifocal | 62 (54.4) | |
|
|
||
| mUICC stage | ||
| I | 13 (11.4) | |
| II | 24 (21.1) | |
| III | 28 (24.6) | |
| IV | 49 (43.0) | |
|
|
||
| AFP (ng/mL) | 112.5 (7–1,412) | 3.8 (2–5) |
|
|
||
| PIVKA-II (mAU/mL) | 159.5 (27–2,431) | 18 (12–23) |
|
|
||
| Exosomal miR-720 | 52.4 (7.8–457.9) | 0.4 (0.2–2.2) |
Values are presented as number (%), median (interquartile range), or mean±standard deviation.
HBV, hepatitis B virus; HCV, hepatitis C virus; AST, aspartate aminotransferase; ALT, alanine aminotransferase; mUICC, modified Union for International Cancer Control; AFP, α-fetoprotein; PIVKA-II, prothrombin-induced by vitamin K absence or antagonist-II.
* The 22 non-HCC control cases include non-viral liver cirrhosis (n=10), alcoholic liver disease (n=7), autoimmune liver disease (n=3), and others (n=2).
- 1. Farinati F, Marino D, De Giorgio M, Baldan A, Cantarini M, Cursaro C, et al. Diagnostic and prognostic role of alpha-fetoprotein in hepatocellular carcinoma: both or neither? Am J Gastroenterol 2006;101:524−532.ArticlePubMed
- 2. Li C, Zhang Z, Zhang P, Liu J. Diagnostic accuracy of des-gamma-carboxy prothrombin versus α-fetoprotein for hepatocellular carcinoma: a systematic review. Hepatol Res 2014;44:E11−E25.ArticlePubMed
- 3. Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, et al. The role of microRNAs in hepatocellular carcinoma. J Cancer 2018;9:3557−3569.ArticlePubMedPMC
- 4. Oura K, Morishita A, Masaki T. Molecular and functional roles of microRNAs in the progression of hepatocellular carcinoma-a review. Int J Mol Sci 2020;21:8362. ArticlePubMedPMC
- 5. Thietart S, Rautou PE. Extracellular vesicles as biomarkers in liver diseases: a clinician’s point of view. J Hepatol 2020;73:1507−1525.ArticlePubMed
- 6. Sharma A, Khatun Z, Shiras A. Tumor exosomes: cellular postmen of cancer diagnosis and personalized therapy. Nanomedicine (Lond) 2016;11:421−437.ArticlePubMed
- 7. Sorop A, Constantinescu D, Cojocaru F, Dinischiotu A, Cucu D, Dima SO. Exosomal microRNAs as biomarkers and therapeutic targets for hepatocellular carcinoma. Int J Mol Sci 2021;22:4997. ArticlePubMedPMC
- 8. Nedaeinia R, Manian M, Jazayeri MH, Ranjbar M, Salehi R, Sharifi M, et al. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther 2017;24:48−56.ArticlePubMed
- 9. Pascut D, Pratama MY, Vo NVT, Masadah R, Tiribelli C. The cross-talk between tumor cells and the microenvironment in hepatocellular carcinoma: the role of exosomal microRNAs and their clinical implications. Cancers (Basel) 2020;12:823. ArticlePubMedPMC
- 10. Korean Liver Cancer Association (KLCA); National Cancer Center (NCC). 2018 Korean Liver Cancer Association-National Cancer Center Korea practice guidelines for the management of hepatocellular carcinoma. Korean J Radiol 2019;20:1042−1113.ArticlePubMedPMCPDF
- 11. Kim HS, Kim JS, Park NR, Nam H, Sung PS, Bae SH, et al. Exosomal miR-125b exerts anti-metastatic properties and predicts early metastasis of hepatocellular carcinoma. Front Oncol 2021;11:637247. ArticlePubMedPMC
- 12. Colli A, Fraquelli M, Casazza G, Massironi S, Colucci A, Conte D, et al. Accuracy of ultrasonography, spiral CT, magnetic resonance, and alpha-fetoprotein in diagnosing hepatocellular carcinoma: a systematic review. Am J Gastroenterol 2006;101:513−523.ArticlePubMed
- 13. Wang T, Zhang KH. New blood biomarkers for the diagnosis of AFP-negative hepatocellular carcinoma. Front Oncol 2020;10:1316. ArticlePubMedPMC
- 14. Liu Z, Pu Y, Bao Y, He S. Investigation of potential molecular biomarkers for diagnosis and prognosis of AFP-negative HCC. Int J Gen Med 2021;14:4369−4380.ArticlePubMedPMC
- 15. Kang KH, Kim JH, Kang SH, Lee BJ, Seo YS, Yim HJ, et al. The influence of alcoholic liver disease on serum PIVKA-II levels in patients without hepatocellular carcinoma. Gut Liver 2015;9:224−230.ArticlePubMed
- 16. Schopman NC, Heynen S, Haasnoot J, Berkhout B. A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol 2010;7:573−576.ArticlePubMed
- 17. Bhat NS, Colden M, Dar AA, Saini S, Arora P, Shahryari V, et al. MicroRNA-720 regulates E-cadherin-αE-catenin complex and promotes renal cell carcinoma. Mol Cancer Ther 2017;16:2840−2848.ArticlePubMedPMC
- 18. Wang X, Kuang Y, Shen X, Zhou H, Chen Y, Han Y, et al. Evaluation of miR-720 prognostic significance in patients with colorectal cancer. Tumour Biol 2015;36:719−727.ArticlePubMed
- 19. Das SG, Romagnoli M, Mineva ND, Barillé-Nion S, Jézéquel P, Campone M, et al. miR-720 is a downstream target of an ADAM8-induced ERK signaling cascade that promotes the migratory and invasive phenotype of triple-negative breast cancer cells. Breast Cancer Res 2016;18:40. ArticlePubMedPMC
- 20. Liu Y, Jiang K, Zhi T, Xu X. miR-720 is a key regulator of glioma migration and invasion by controlling TARSL2 expression. Hum Cell 2021;34:1504−1516.ArticlePubMed
- 21. Chen P, Zhang G, Zhou Q, Li Z. Plasma microRNA-720 may predict prognosis and diagnosis in glioma patients. Biosci Rep 2020;40:BSR20201449. ArticlePubMedPMC
- 22. Li LZ, Zhang CZ, Liu LL, Yi C, Lu SX, Zhou X, et al. miR-720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis 2014;35:469−478.ArticlePubMed
References
Figure & Data
References
Citations

 PubReader
PubReader ePub Link
ePub Link Download Citation
Download Citation
- Download Citation
- Close
- Related articles
-
- Imaging prognostication and tumor biology in hepatocellular carcinoma
- Non-alcoholic fatty liver disease-related hepatocellular carcinoma
- A clinical and pathological update on hepatocellular carcinoma
- A Case of Metastatic Melanoma in the Liver Mimicking Hepatocellular Carcinoma
- Liver Magnetic Resonance Imaging for Hepatocellular Carcinoma Surveillance

 E-submission
E-submission THE KOREAN LIVER CANCER ASSOCIATION
THE KOREAN LIVER CANCER ASSOCIATION


 Follow JLC on Twitter
Follow JLC on Twitter